Network activity¶
The previous notebook provided us with an anatomical reconstruction of the barrel cortex, defining locations of presynaptic cells, and post-synaptic targets onto our cell. Given this anatomical data, we can now activate the synapses according to experimental data.
To do this, we need the following information
A parameter file specifying characteristics of the synaspses by celltype
A parameterfile specifying the ongoing activity by celltype
Parameterfiles specifying evoked response by stimulus, celltype and cell location
[17]:
from pathlib import Path
tutorial_output_dir = f"{Path.home()}/isf_tutorial_output" # <-- Change this to your desired output directory
[19]:
import Interface as I
%matplotlib inline
db = I.DataBase(tutorial_output_dir)['network_modeling']
con_file = db['anatomical_constraints'].get_file('.con')
con_file_path = db['anatomical_constraints'].join(con_file)
syn_file = db['anatomical_constraints'].get_file('.syn')
syn_file_path = db['anatomical_constraints'].join(syn_file)
Characterizing ongoing activity¶
[20]:
from getting_started import getting_started_dir
ongoing_template_param_name = I.os.path.join(
getting_started_dir,
'example_data',
'functional_constraints',
'ongoing_activity',
'ongoing_activity_celltype_template_exc_conductances_fitted.param')
In this parameter file, the following keys are defined
[21]:
ongoing_template_param = I.scp.build_parameters(ongoing_template_param_name)
ongoing_template_param.keys()
[21]:
dict_keys(['info', 'network', 'NMODL_mechanisms'])
The most relevant information is specified in the network key:
[22]:
ongoing_template_param.network.keys()
[22]:
dict_keys(['L2', 'L34', 'L4py', 'L4sp', 'L4ss', 'L5st', 'L5tt', 'L6cc', 'L6ccinv', 'L6ct', 'VPM', 'L1', 'L23Trans', 'L45Peak', 'L45Sym', 'L56Trans', 'SymLocal1', 'SymLocal2', 'SymLocal3', 'SymLocal4', 'SymLocal5', 'SymLocal6'])
Here, parameters are defined for each presynaptic celltype. Let’s inspect the L5tt parameters for example:
[23]:
ongoing_template_param.network.L5tt
[23]:
NTParameterSet({'celltype': 'spiketrain', 'interval': 283.3, 'synapses': NTParameterSet({'receptors': NTParameterSet({'glutamate_syn': NTParameterSet({'threshold': 0.0, 'delay': 0.0, 'parameter': NTParameterSet({'tau1': 26.0, 'tau2': 2.0, 'tau3': 2.0, 'tau4': 0.1, 'decayampa': 1.0, 'decaynmda': 1.0, 'facilampa': 0.0, 'facilnmda': 0.0}), 'weight': [1.59, 1.59]})}), 'releaseProb': 0.6})})
interval: mean ongoing interspike intervalreleaseProb: chance that synapse gets activated if presynaptic cell gets activatedsynapses.receptors.glutamate_syn.parameter:parameters for the NEURON mechanism defined in mechanisms.channels
tau1: NMDA decay timetau2: NMDA rise timetau3: AMPA decay timetau4: AMPA rise time
synapses.receptors.weightmaximum conductance of the synapse for AMPA and NMDA, respectively
Characterizing evoked activity¶
Evoked activity captures the network activity during a specific in vivo condition. The experimental condition we will consider here is a passive whisker touch of the rat. We have files capturing the activity of all celltypes in all locations across the rat barrel cortex. Let’s see what their activity was when we touched the whisker in arc C, row 2.
[24]:
evokedPrefix = I.os.path.join(getting_started_dir, 'example_data/functional_constraints/evoked_activity/')
excitatory_PSTHs = [fname for fname in I.os.listdir(evokedPrefix) if fname.endswith('PSTH_UpState.param')]
inhibitory_PSTHs = [fname for fname in I.os.listdir(evokedPrefix) if fname.endswith('active_timing_normalized_PW_1.0_SuW_0.5.param')]
Let’s have a look at a specific parameterfile:
[25]:
excitatory_PSTHs
[25]:
['L4ss_3x3_PSTH_UpState.param',
'L4sp_3x3_PSTH_UpState.param',
'L4py_3x3_PSTH_UpState.param',
'L5st_3x3_PSTH_UpState.param',
'L6ct_3x3_PSTH_UpState.param',
'L6cc_3x3_PSTH_UpState.param',
'L6ccinv_3x3_PSTH_UpState.param',
'L2_3x3_PSTH_UpState.param',
'L34_3x3_PSTH_UpState.param',
'L5tt_3x3_PSTH_UpState.param']
These files simply define post-stimulus time histograms (PSTH) for varying cell types, relative to a stimulus onset. For more information on the file format of this activity data, you can visit the documentation page on activity data.
[26]:
example_PSTH_L6cc = I.scp.build_parameters(I.os.path.join(evokedPrefix, 'L6cc_3x3_PSTH_UpState.param'))
example_PSTH_L6cc.keys()
[26]:
dict_keys(['L6cc_B1', 'L6cc_B2', 'L6cc_B3', 'L6cc_C1', 'L6cc_C2', 'L6cc_C3', 'L6cc_D1', 'L6cc_D2', 'L6cc_D3'])
We have entries for each column, each containing the respective evoked PSTH in a C2 stimulus scenario:
[27]:
example_PSTH_L6cc
[27]:
NTParameterSet({'L6cc_B1': NTParameterSet({'distribution': 'PSTH', 'intervals': [[10.0, 11.0], [11.0, 12.0], [20.0, 21.0], [22.0, 23.0], [32.0, 33.0], [33.0, 34.0], [34.0, 35.0], [36.0, 37.0], [37.0, 38.0], [41.0, 42.0], [43.0, 44.0], [45.0, 46.0]], 'probabilities': [0.0034, 0.0034, 0.0072, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034]}), 'L6cc_B2': NTParameterSet({'distribution': 'PSTH', 'intervals': [[10.0, 11.0], [17.0, 18.0], [20.0, 21.0], [25.0, 26.0], [34.0, 35.0], [36.0, 37.0], [37.0, 38.0], [41.0, 42.0]], 'probabilities': [0.0034, 0.0034, 0.0072, 0.0034, 0.0034, 0.0034, 0.0034, 0.0072]}), 'L6cc_B3': NTParameterSet({'distribution': 'PSTH', 'intervals': [[5.0, 6.0], [7.0, 8.0], [16.0, 17.0], [23.0, 24.0], [28.0, 29.0], [38.0, 39.0], [40.0, 41.0], [44.0, 45.0], [46.0, 47.0], [49.0, 50.0]], 'probabilities': [0.0034, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034, 0.0072, 0.0034, 0.0034, 0.0034]}), 'L6cc_C1': NTParameterSet({'distribution': 'PSTH', 'intervals': [[7.0, 8.0], [8.0, 9.0], [9.0, 10.0], [10.0, 11.0], [11.0, 12.0], [12.0, 13.0], [13.0, 14.0], [16.0, 17.0], [18.0, 19.0], [20.0, 21.0], [27.0, 28.0], [31.0, 32.0], [33.0, 34.0], [34.0, 35.0], [36.0, 37.0], [38.0, 39.0], [40.0, 41.0], [42.0, 43.0], [48.0, 49.0]], 'probabilities': [0.0034, 0.0034, 0.0148, 0.0681, 0.0377, 0.011, 0.0034, 0.0072, 0.0072, 0.0034, 0.0034, 0.0034, 0.0072, 0.0072, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034]}), 'L6cc_C2': NTParameterSet({'distribution': 'PSTH', 'intervals': [[3.0, 4.0], [6.0, 7.0], [9.0, 10.0], [10.0, 11.0], [11.0, 12.0], [12.0, 13.0], [13.0, 14.0], [14.0, 15.0], [15.0, 16.0], [18.0, 19.0], [19.0, 20.0], [33.0, 34.0], [34.0, 35.0], [35.0, 36.0], [36.0, 37.0], [41.0, 42.0], [42.0, 43.0], [43.0, 44.0]], 'probabilities': [0.0034, 0.0034, 0.0186, 0.0757, 0.0491, 0.0186, 0.0034, 0.0034, 0.0072, 0.0034, 0.0034, 0.0072, 0.0034, 0.0072, 0.0034, 0.0034, 0.0072, 0.0034]}), 'L6cc_C3': NTParameterSet({'distribution': 'PSTH', 'intervals': [[4.0, 5.0], [9.0, 10.0], [10.0, 11.0], [14.0, 15.0], [15.0, 16.0], [16.0, 17.0], [17.0, 18.0], [21.0, 22.0], [22.0, 23.0], [26.0, 27.0], [31.0, 32.0], [40.0, 41.0], [47.0, 48.0], [48.0, 49.0]], 'probabilities': [0.0034, 0.0034, 0.011, 0.0072, 0.0034, 0.011, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034, 0.0034]}), 'L6cc_D1': NTParameterSet({'distribution': 'PSTH', 'intervals': [[25.0, 26.0], [27.0, 28.0], [37.0, 38.0], [43.0, 44.0]], 'probabilities': [0.0034, 0.0034, 0.0034, 0.0034]}), 'L6cc_D2': NTParameterSet({'distribution': 'PSTH', 'intervals': [[9.0, 10.0], [10.0, 11.0], [30.0, 31.0], [37.0, 38.0], [41.0, 42.0], [42.0, 43.0], [43.0, 44.0], [45.0, 46.0], [48.0, 49.0]], 'probabilities': [0.0034, 0.0034, 0.0034, 0.0072, 0.0034, 0.0034, 0.0034, 0.0034, 0.0072]}), 'L6cc_D3': NTParameterSet({'distribution': 'PSTH', 'intervals': [[1.0, 2.0], [11.0, 12.0], [31.0, 32.0], [49.0, 50.0]], 'probabilities': [0.0041, 0.0041, 0.0041, 0.0041]})})
Let’s see what these PSTHs actually look like:
[28]:
%matplotlib inline
I.plt.style.use("fivethirtyeight")
rows, cols = (1, 2, 3), ("B", "C", "D")
fig, axs = I.plt.subplots(len(rows), len(cols), sharey=True, sharex=True, figsize=(10, 6))
for cell_type, psth in example_PSTH_L6cc.items():
p, bins = psth['probabilities'], psth['intervals']
barrel_ind, layer = cell_type.split('_')[-1]
ax_row_ind, ax_col_ind = rows.index(int(layer)), cols.index(barrel_ind)
for b, p_ in zip(bins, p):
axs[ax_row_ind, ax_col_ind].bar(b, p_, color="C0")
if ax_col_ind == 0: axs[ax_row_ind, ax_col_ind].set_ylabel("Arc {}".format(rows[ax_row_ind]))
if ax_row_ind == 0: axs[ax_row_ind, ax_col_ind].set_title("Row {}".format(cols[ax_col_ind]))
I.plt.suptitle("Response probability of L6cc neurons in barrel C2\nto deflecting whiskers arcs [1, 2, 3] x rows [B, C, D]")
fig.tight_layout()
I.plt.show()
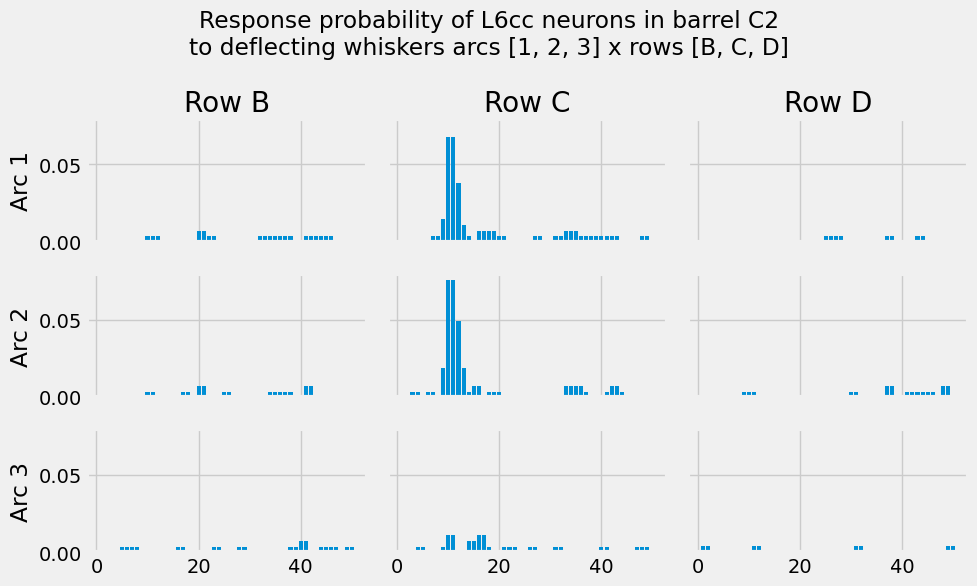
Combining ongoing and evoked activity into a network parameter file¶
Now, let’s reverse-engineer our network parameters from this. Given that we know how a single cell population responds to various input stimuli, we can now generate activity data for all cell populations under a single stimulus.
To so, we use the create_evoked_network_parameter pipeline. This pipeline has been built specifically for reading in Activity data organized per barrel cortex column. For more information on this data, please refer to:
The actual data under
getting_started/example_data/functional_constraints/evoked-activityevoked_network_param_from_template for an example on how these files are read and parsed, although this is also covered below.
One relevant detail of this pipeline is that the stimulus is set to come in at \(245 ms\) by default. This can later be seen by checking the offset parameter in the Network parameters for each cell type. Keep this in mind when inspecting PSTHs that relate to evoked activity: they start at \(0ms\), but during simulations, they start at offset.
[29]:
whisker = 'C2'
# cell number spreadsheet generated in anatomical realization step.
cell_number_file_name = db['anatomical_constraints'].join('NumberOfConnectedCells.csv')
if not 'network_param' in db.keys(): db.create_managed_folder('network_param')
out_file_name = db['network_param'].join('C2_stim.param')
I.create_evoked_network_parameter(
ongoing_template_param_name,
cell_number_file_name,
syn_file_path,
con_file_path,
whisker,
out_file_name
)
*************
creating network parameter file from template /gpfs/soma_fs/scratch/meulemeester/project_src/in_silico_framework/getting_started/example_data/functional_constraints/ongoing_activity/ongoing_activity_celltype_template_exc_conductances_fitted.param
*************
Let’s also generate parameterfiles for surround whisker stimuli! These will be used in the next tutorial to run multi-scale simulations.
[ ]:
surround_whiskers = ['B1', 'B2', 'B3', 'C1', 'C3', 'D1', 'D2', 'D3']
with I.silence_stdout:
for whisker in surround_whiskers:
I.create_evoked_network_parameter(
ongoing_template_param_name,
cell_number_file_name,
syn_file_path, con_file_path,
whisker,
db['network_param'].join('{}_stim.param'.format(whisker))
)
And one more time to create parameterfiles for \(2nd\) surround stimuli. These will be used in Tutorial 4.2 to manipulate synaptic input
[31]:
second_surround_whiskers = [
'A1', 'A2', 'A3', 'A4',
'E1', 'E2', 'E3', 'E4',
'B4', 'C4', 'D4'
]
with I.silence_stdout:
for whisker in second_surround_whiskers:
I.create_evoked_network_parameter(
ongoing_template_param_name,
cell_number_file_name,
syn_file_path, con_file_path,
whisker,
db['network_param'].join('{}_stim.param'.format(whisker))
)
The resulting network parameters describe ongoing and evoked activity for each synapse in a cell type specific manner.
Inspecting the network activity¶
Let’s visualize the response probabilities of each cell type, similar to what we did above.
[32]:
network_param_C2 = I.scp.build_parameters(db['network_param'].join('C2_stim.param'))
network_param_C2.network.keys()
[32]:
dict_keys(['L2_Alpha', 'L2_B1', 'L2_B2', 'L2_B3', 'L2_B4', 'L2_Beta', 'L2_C1', 'L2_C2', 'L2_C3', 'L2_C4', 'L2_D1', 'L2_D2', 'L2_D3', 'L2_D4', 'L2_E1', 'L2_E2', 'L2_Gamma', 'L34_A1', 'L34_A2', 'L34_A3', 'L34_A4', 'L34_Alpha', 'L34_B1', 'L34_B2', 'L34_B3', 'L34_B4', 'L34_Beta', 'L34_C1', 'L34_C2', 'L34_C3', 'L34_C4', 'L34_D1', 'L34_D2', 'L34_D3', 'L34_Delta', 'L34_E1', 'L34_E2', 'L34_E3', 'L34_Gamma', 'L4py_A1', 'L4py_A2', 'L4py_A3', 'L4py_A4', 'L4py_Alpha', 'L4py_B1', 'L4py_B2', 'L4py_B3', 'L4py_B4', 'L4py_C1', 'L4py_C2', 'L4py_C3', 'L4py_C4', 'L4py_D1', 'L4py_D2', 'L4py_D3', 'L4py_Gamma', 'L4sp_B1', 'L4sp_B2', 'L4sp_B3', 'L4sp_Beta', 'L4sp_C1', 'L4sp_C2', 'L4sp_C3', 'L4sp_C4', 'L4sp_D1', 'L4sp_D2', 'L4sp_D3', 'L4sp_Delta', 'L4sp_Gamma', 'L4ss_A1', 'L4ss_Alpha', 'L4ss_B1', 'L4ss_B2', 'L4ss_B3', 'L4ss_C1', 'L4ss_C2', 'L4ss_C3', 'L4ss_D1', 'L4ss_D2', 'L4ss_D3', 'L5st_A1', 'L5st_A2', 'L5st_A3', 'L5st_Alpha', 'L5st_B1', 'L5st_B2', 'L5st_B3', 'L5st_B4', 'L5st_Beta', 'L5st_C1', 'L5st_C2', 'L5st_C3', 'L5st_C4', 'L5st_D1', 'L5st_D2', 'L5st_D3', 'L5st_Delta', 'L5st_E1', 'L5st_Gamma', 'L5tt_A1', 'L5tt_A3', 'L5tt_A4', 'L5tt_B1', 'L5tt_B2', 'L5tt_B3', 'L5tt_B4', 'L5tt_Beta', 'L5tt_C1', 'L5tt_C2', 'L5tt_C3', 'L5tt_C4', 'L5tt_D2', 'L5tt_D3', 'L5tt_E1', 'L6cc_A1', 'L6cc_A2', 'L6cc_A3', 'L6cc_A4', 'L6cc_Alpha', 'L6cc_B1', 'L6cc_B2', 'L6cc_B3', 'L6cc_B4', 'L6cc_Beta', 'L6cc_C1', 'L6cc_C2', 'L6cc_C3', 'L6cc_C4', 'L6cc_D1', 'L6cc_D2', 'L6cc_D3', 'L6cc_D4', 'L6cc_E1', 'L6cc_E2', 'L6cc_E3', 'L6cc_E4', 'L6cc_Gamma', 'L6ccinv_A2', 'L6ccinv_A3', 'L6ccinv_A4', 'L6ccinv_Alpha', 'L6ccinv_B1', 'L6ccinv_B2', 'L6ccinv_B3', 'L6ccinv_B4', 'L6ccinv_Beta', 'L6ccinv_C1', 'L6ccinv_C2', 'L6ccinv_C3', 'L6ccinv_C4', 'L6ccinv_D1', 'L6ccinv_D2', 'L6ccinv_D3', 'L6ccinv_D4', 'L6ccinv_E1', 'L6ccinv_E2', 'L6ccinv_E3', 'L6ccinv_E4', 'L6ccinv_Gamma', 'L6ct_Alpha', 'L6ct_B1', 'L6ct_B2', 'L6ct_B3', 'L6ct_B4', 'L6ct_Beta', 'L6ct_C1', 'L6ct_C2', 'L6ct_C3', 'L6ct_D1', 'L6ct_D2', 'L6ct_D3', 'L6ct_E3', 'L6ct_Gamma', 'VPM_B1', 'VPM_B2', 'VPM_B3', 'VPM_C1', 'VPM_C2', 'VPM_C3', 'VPM_D1', 'VPM_D2', 'VPM_Gamma', 'L1_B1', 'L1_B2', 'L1_B3', 'L1_Beta', 'L1_C1', 'L1_C2', 'L1_C3', 'L1_D1', 'L1_D2', 'L1_D3', 'L1_E1', 'L1_E2', 'L23Trans_B2', 'L23Trans_B3', 'L23Trans_C1', 'L23Trans_C2', 'L23Trans_C3', 'L23Trans_D1', 'L23Trans_D2', 'L45Peak_B1', 'L45Peak_B2', 'L45Peak_B3', 'L45Peak_B4', 'L45Peak_C1', 'L45Peak_C2', 'L45Peak_C3', 'L45Peak_C4', 'L45Peak_D1', 'L45Peak_D2', 'L45Peak_D3', 'L45Peak_Delta', 'L45Peak_E1', 'L45Sym_A1', 'L45Sym_A2', 'L45Sym_B1', 'L45Sym_B2', 'L45Sym_B3', 'L45Sym_B4', 'L45Sym_Beta', 'L45Sym_C1', 'L45Sym_C2', 'L45Sym_C3', 'L45Sym_D1', 'L45Sym_D2', 'L45Sym_D3', 'L56Trans_A1', 'L56Trans_A2', 'L56Trans_A3', 'L56Trans_A4', 'L56Trans_B1', 'L56Trans_B2', 'L56Trans_B3', 'L56Trans_B4', 'L56Trans_C1', 'L56Trans_C2', 'L56Trans_C3', 'L56Trans_D1', 'L56Trans_D2', 'L56Trans_D3', 'L56Trans_E1', 'L56Trans_Gamma', 'SymLocal1_B2', 'SymLocal1_C1', 'SymLocal1_C2', 'SymLocal1_C3', 'SymLocal1_D1', 'SymLocal1_D2', 'SymLocal2_B2', 'SymLocal2_B3', 'SymLocal2_C2', 'SymLocal3_B2', 'SymLocal3_B3', 'SymLocal3_B4', 'SymLocal3_C1', 'SymLocal3_C2', 'SymLocal3_C3', 'SymLocal3_D1', 'SymLocal3_D2', 'SymLocal4_A2', 'SymLocal4_Alpha', 'SymLocal4_B1', 'SymLocal4_B2', 'SymLocal4_B3', 'SymLocal4_B4', 'SymLocal4_C1', 'SymLocal4_C2', 'SymLocal4_C3', 'SymLocal4_D1', 'SymLocal4_D2', 'SymLocal4_Delta', 'SymLocal4_E1', 'SymLocal4_Gamma', 'SymLocal5_A3', 'SymLocal5_A4', 'SymLocal5_B2', 'SymLocal5_B3', 'SymLocal5_C2', 'SymLocal5_D2', 'SymLocal6_C2', 'SymLocal6_D1'])
[33]:
surround_column_map = {
'A1': {'Alpha': 4, 'A1': 5, 'A2': 6, 'B1': 8, 'B2': 9},\
'A2': {'A1': 4, 'A2': 5, 'A3': 6, 'B1': 7, 'B2': 8, 'B3': 9},\
'A3': {'A2': 4, 'A3': 5, 'A4': 6, 'B2': 7, 'B3': 8, 'B4': 9},\
'A4': {'A3': 4, 'A4': 5, 'B3': 7, 'B4': 8},\
'Alpha': {'Alpha': 5, 'A1': 6, 'Beta': 8, 'B1': 9},\
'B1': {'Alpha': 1, 'A1': 2, 'A2': 3, 'Beta': 4, 'B1': 5, 'B2': 6, 'C1': 8, 'C2': 9},\
'B2': {'A1': 1, 'A2': 2, 'A3': 3, 'B1': 4, 'B2': 5, 'B3': 6, 'C1': 7, 'C2': 8, 'C3': 9},\
'B3': {'A2': 1, 'A3': 2, 'A4': 3, 'B2': 4, 'B3': 5, 'B4': 6, 'C2': 7, 'C3': 8, 'C4': 9},\
'B4': {'A3': 1, 'A4': 2, 'B3': 4, 'B4': 5, 'C3': 7, 'C4': 8},\
'Beta': {'Alpha': 2, 'Beta': 5, 'B1': 6, 'Gamma': 8, 'C1': 9},\
'C1': {'Beta': 1, 'B1': 2, 'B2': 3, 'Gamma': 4, 'C1': 5, 'C2': 6, 'D1': 8, 'D2': 9},\
'C2': {'B1': 1, 'B2': 2, 'B3': 3, 'C1': 4, 'C2': 5, 'C3': 6, 'D1': 7, 'D2': 8, 'D3': 9},\
'C3': {'B2': 1, 'B3': 2, 'B4': 3, 'C2': 4, 'C3': 5, 'C4': 6, 'D2': 7, 'D3': 8, 'D4': 9},\
'C4': {'B3': 1, 'B4': 2, 'C3': 4, 'C4': 5, 'D3': 7, 'D4': 8},\
'Gamma': {'Beta': 2, 'Gamma': 5, 'C1': 6, 'Delta': 8, 'D1': 9},\
'D1': {'Gamma': 1, 'C1': 2, 'C2': 3, 'Delta': 4, 'D1': 5, 'D2': 6, 'E1': 8, 'E2': 9},\
'D2': {'C1': 1, 'C2': 2, 'C3': 3, 'D1': 4, 'D2': 5, 'D3': 6, 'E1': 7, 'E2': 8, 'E3': 9},\
'D3': {'C2': 1, 'C3': 2, 'C4': 3, 'D2': 4, 'D3': 5, 'D4': 6, 'E2': 7, 'E3': 8, 'E4': 9},\
'D4': {'C3': 1, 'C4': 2, 'D3': 4, 'D4': 5, 'E3': 7, 'E4': 8},\
'Delta': {'Gamma': 2, 'Delta': 5, 'D1': 6, 'E1': 9},\
'E1': {'Delta': 1, 'D1': 2, 'D2': 3, 'E1': 5, 'E2': 6},\
'E2': {'D1': 1, 'D2': 2, 'D3': 3, 'E1': 4, 'E2': 5, 'E3': 6},\
'E3': {'D2': 1, 'D3': 2, 'D4': 3, 'E2': 4, 'E3': 5, 'E4': 6},\
'E4': {'D3': 1, 'D4': 2, 'E3': 4, 'E4': 5}}
def activity_gridplot(ct, deflected_whisker="C2", network_param=network_param_C2):
surround_columns = surround_column_map[deflected_whisker]
rows = list(set([e[1] for e in surround_columns]))
n_rows = len(rows)
columns = list(set([e[0] for e in surround_columns]))
columns.sort()
n_columns = len(columns)
fig, axs = I.plt.subplots(n_rows, n_columns, sharey=True, sharex=True)
for surround_column in surround_columns:
row = int(surround_column[-1])
column = surround_column[-2]
axs[row-1][0].set_ylabel(row)
axs[0][columns.index(column)].set_xlabel(column)
axs[0][columns.index(column)].xaxis.set_label_position('top')
cell_type = ct+"_{}{}".format(column, row)
if cell_type not in network_param.network.keys():
continue
if not 'pointcell' in network_param.network[cell_type]['celltype']:
continue
x = [b[0] for b in network_param.network[cell_type]['celltype']['pointcell']['intervals']]
if row == 2 and column == "C":
# barrel of the whisker deflection
color = "C1"
else:
color = "Grey"
bar = axs[row-1][columns.index(column)].bar(
x,
network_param.network[cell_type]['celltype']['pointcell']['probabilities'],
facecolor=color,
edgecolor=color,
lw=2)
fig.supylabel("Barrel cortex row")
fig.suptitle(
"Response probability of {} populations\nwhen deflecting whisker {}".format(
ct, deflected_whisker))
fig.tight_layout()
return fig, axs
[34]:
ct = "L6cc"
fig, axs = activity_gridplot(ct)
I.plt.show()
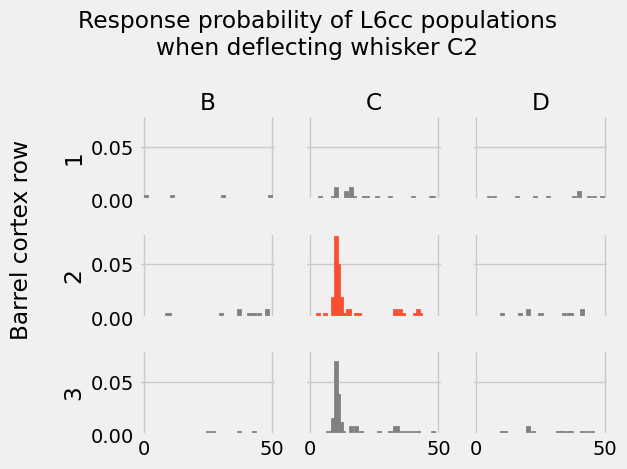
Note again that this time we are not visualizing the response probability of some neuron population to different inputs (like the first histogram grid). We are instead visualizing the response probabilities of different neuron populations to the same experimental condition: deflecting whisker C2.
[18]:
ct = "L5tt"
fig, axs = activity_gridplot(ct)
I.plt.show()
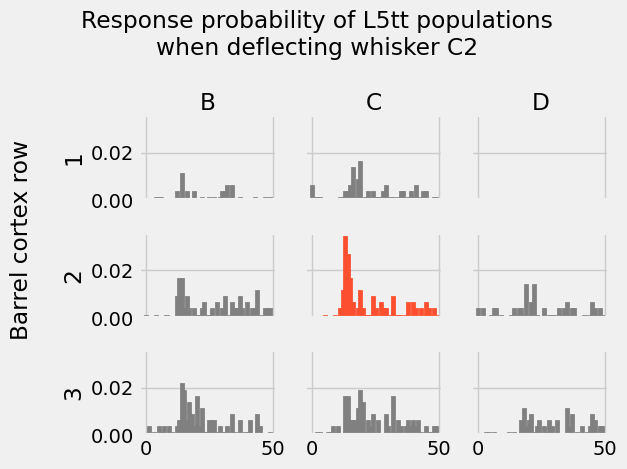
Recap¶
Tutorial 2.1 covered how to embed a neuron model into a network reconstruction, taking into account the relevant morphological, cellular and network properties. This tutorial covered how to generate synaptic activity. We are now ready to combine our biophysically detailed neuron model with this network model: see Tutorial 3.1.