Welcome to the In Silico Framework (ISF)¶
ISF is a multi-scale simulation environment for the generation, simulation, and analysis of neurobiologically tractable single cell and network-level simulations.
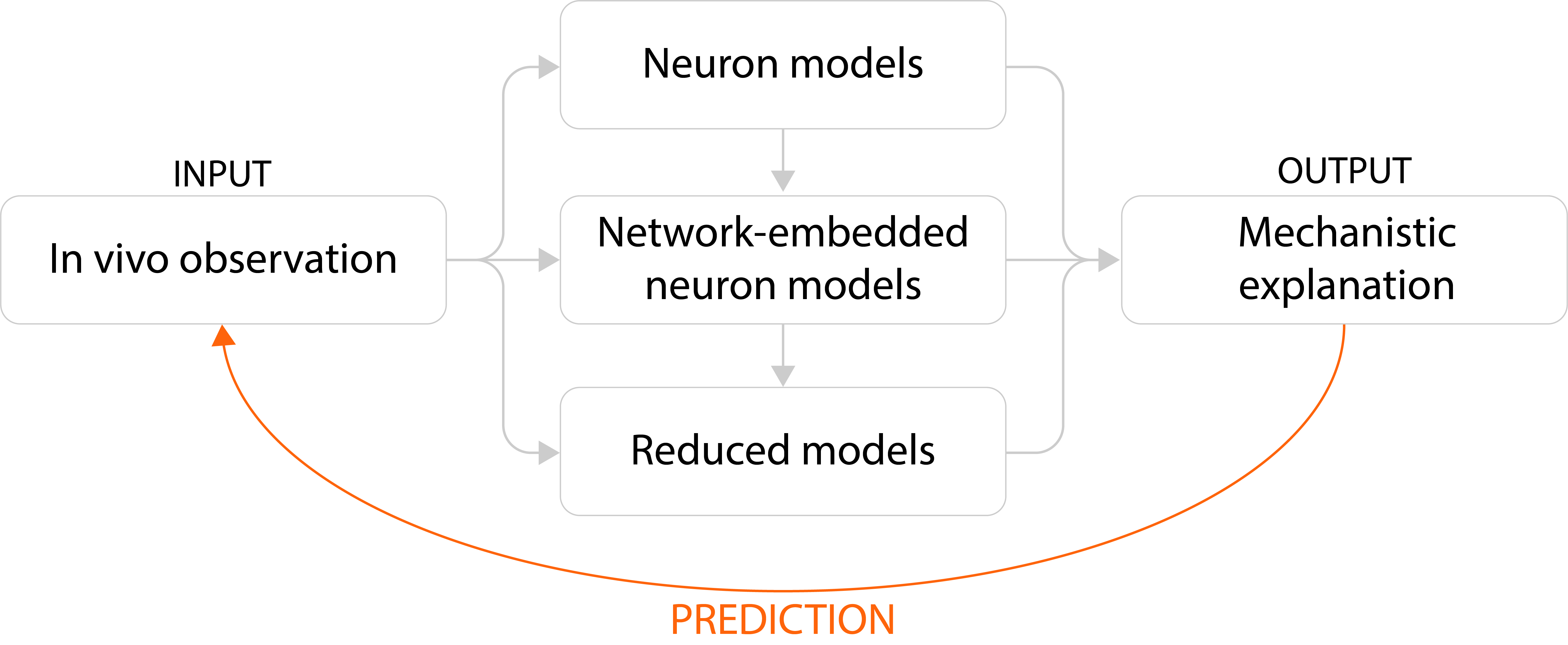
Packages in ISF¶
Packages in this repository you will most likely directly interact with¶
Module |
Description |
|---|---|
|
Module for generating anatomical models, i.e. determining number and location of synapses and number and location of presynaptic cells |
|
High level interface to the NEURON simulator providing methods to perform single cell simulations with synaptic input. The anatomical constraints of the synaptic input are provided by the single_cell_input_mapper module. |
|
High level interface to the single_cell_parser module providing methods for common simulation tasks. It also provides methods for building reduced models mimicing the full compartmental model. |
|
Flexible database whose API mimics a python dictionary. It provides efficient and scalable methods to store and access simulation results at a terrabyte scale. It also generates metadata, indicating when the data was put in the database and the exact version of the in_silico_framework that was used at this timepoint. Simulation results from the single_cell_parser module can be imported and converted to a high performance binary format. |
How should I interact with these packages?¶
The recommended way is to use the Interface module which provides the API necessary to perform simulation tasks.
[1]:
import Interface as I
[INFO] ISF: Current version: heads/develop
[INFO] ISF: Current pid: 39666
[INFO] ISF: Loading mechanisms:
Warning: no DISPLAY environment variable.
--No graphics will be displayed.
[INFO] ISF: Loaded modules with __version__ attribute are:
IPython: 8.12.2, Interface: heads/develop, PIL: 10.4.0, _brotli: 1.0.9, _csv: 1.0, _ctypes: 1.1.0, _curses: b'2.2', _decimal: 1.70, argparse: 1.1, backcall: 0.2.0, blosc: 1.11.1, bluepyopt: 1.9.126, brotli: 1.0.9, certifi: 2024.08.30, cffi: 1.17.0, charset_normalizer: 3.4.0, click: 7.1.2, cloudpickle: 3.1.0, colorama: 0.4.6, comm: 0.2.2, csv: 1.0, ctypes: 1.1.0, cycler: 0.12.1, cytoolz: 0.12.3, dash: 2.18.2, dask: 2.30.0, dateutil: 2.9.0, deap: 1.4, debugpy: 1.8.5, decimal: 1.70, decorator: 5.1.1, defusedxml: 0.7.1, distributed: 2.30.0, distutils: 3.8.20, django: 1.8.19, entrypoints: 0.4, executing: 2.1.0, fasteners: 0.17.3, flask: 1.1.4, fsspec: 2024.10.0, future: 1.0.0, greenlet: 3.1.1, idna: 3.10, ipaddress: 1.0, ipykernel: 6.29.5, ipywidgets: 8.1.5, isf_pandas_msgpack: 0.2.2, itsdangerous: 1.1.0, jedi: 0.19.1, jinja2: 2.11.3, joblib: 1.4.2, json: 2.0.9, jupyter_client: 7.3.4, jupyter_core: 5.7.2, kiwisolver: 1.4.5, logging: 0.5.1.2, markupsafe: 2.0.1, matplotlib: 3.5.1, msgpack: 1.0.8, neuron: 7.8.2+, numcodecs: 0.12.1, numexpr: 2.8.6, numpy: 1.19.2, packaging: 24.2, pandas: 1.1.3, parameters: 0.2.1, parso: 0.8.4, past: 1.0.0, pexpect: 4.9.0, pickleshare: 0.7.5, platform: 1.0.8, platformdirs: 4.3.6, plotly: 5.24.1, prompt_toolkit: 3.0.48, psutil: 6.0.0, ptyprocess: 0.7.0, pure_eval: 0.2.3, pydevd: 2.9.5, pygments: 2.18.0, pyparsing: 3.1.4, pytz: 2024.2, re: 2.2.1, requests: 2.32.3, scandir: 1.10.0, scipy: 1.5.2, seaborn: 0.12.2, six: 1.16.0, sklearn: 0.23.2, socketserver: 0.4, socks: 1.7.1, sortedcontainers: 2.4.0, stack_data: 0.6.2, statsmodels: 0.13.2, sumatra: 0.7.4, tables: 3.8.0, tblib: 3.0.0, tlz: 0.12.3, toolz: 1.0.0, traitlets: 5.14.3, urllib3: 2.2.3, wcwidth: 0.2.13, werkzeug: 1.0.1, yaml: 5.3.1, zarr: 2.15.0, zlib: 1.0, zmq: 26.2.0, zstandard: 0.19.0
Now, you can access the relevant packages, functions and classes directly:
[2]:
I.scp # single_cell_parser package
I.sca # single_cell_analyzer package
I.DataBase # main class of isf_data_base
I.map_singlecell_inputs # compute anatomical model for a given cell morphology in barrel cortex
I.simrun_run_new_simulations # default function for running single cell simulations with well constrained synaptic input
I.db_init_simrun_general.init # default method to initialize a model data base with existing simulation results
I.db_init_simrun_general.optimize # converts the data to speed optimized compressed binary format
I.synapse_activation_binning_dask # parallelized binning of synapse activation data
I.rm_get_kernel # create reduced lda model from simulation data
I.silence_stdout # context manager and decorator to silence functions
I.cache # decorator to cache functions
I.np # numpy
I.pd # pandas
I.dask # dask
I.distributed # distributed
I.sns # seaborn
# ...
[2]:
<module 'seaborn' from '/gpfs/soma_fs/scratch/meulemeester/project_src/in_silico_framework/.pixi/envs/default/lib/python3.8/site-packages/seaborn/__init__.py'>
Use autocompletion of IPython to find the other modules. To quickly inspect the documentation from within the IPython or Jupyter session, use a questionmark:
[3]:
I.db_init_simrun_general.init?
Signature:
I.db_init_simrun_general.init(
db,
simresult_path,
core=True,
voltage_traces=True,
synapse_activation=True,
dendritic_voltage_traces=True,
parameterfiles=True,
spike_times=True,
burst_times=False,
repartition=True,
scheduler=None,
rewrite_in_optimized_format=True,
dendritic_spike_times=True,
dendritic_spike_times_threshold=-30.0,
client=None,
n_chunks=5000,
dumper=<module 'data_base.isf_data_base.IO.LoaderDumper.pandas_to_msgpack' from '/gpfs/soma_fs/scratch/meulemeester/project_src/in_silico_framework/data_base/isf_data_base/IO/LoaderDumper/pandas_to_msgpack.py'>,
)
Docstring:
Initialize a database with simulation data.
Use this function to load simulation data generated with the simrun module
into a :py:class:`~data_base.isf_data_base.isf_data_base.ISFDataBase`.
Args:
core (bool, optional):
Parse and write the core data to the database: voltage traces, metadata, sim_trial_index and filelist.
See also: :py:meth:`~data_base.db_initializers.load_simrun_general._build_core`
voltage_traces (bool, optional):
Parse and write the somatic voltage traces to the database.
spike_times (bool, optional):
Parse and write the spike times into the database.
See also: :py:meth:`data_base.analyze.spike_detection.spike_detection`
dendritic_voltage_traces (bool, optional):
Parse and write the dendritic voltage traces to the database.
See also: :py:meth:`~data_base.db_initializers.load_simrun_general._build_dendritic_voltage_traces`
dendritic_spike_times (bool, optional):
Parse and write the dendritic spike times to the database.
See also: :py:meth:`~data_base.db_initializers.load_simrun_general.add_dendritic_spike_times`
dendritic_spike_times_threshold (float, optional):
Threshold for the dendritic spike times in mV. Default is :math:`-30`.
See also: :py:meth:`~data_base.db_initializers.load_simrun_general.add_dendritic_spike_times`
synapse_activation (bool, optional):
Parse and write the synapse activation data to the database.
See also: :py:meth:`~data_base.db_initializers.load_simrun_general._build_synapse_activation`
parameterfiles (bool, optional):
Parse and write the parameterfiles to the database.
See also: :py:meth:`~data_base.db_initializers.load_simrun_general._build_param_files`
rewrite_in_optimized_format (bool, optional):
If True (default): data is converted to a high performance binary
format and makes unpickling more robust against version changes of third party libraries.
Also, it makes the database self-containing, i.e. you can move it to another machine or
subfolder and everything still works. Deleting the data folder then would (should) not cause
loss of data.
If False: the db only contains links to the actual simulation data folder
and will not work if the data folder is deleted or moved or transferred to another machine
where the same absolute paths are not valid.
repartition (bool, optional):
If True, the dask dataframe is repartitioned to 5000 partitions (only if it contains over :math:`10000` entries).
n_chunks (int, optional):
Number of chunks to split the :ref:`syn_activation_format` and :ref:`spike_times_format` dataframes into.
Default is 5000.
client (dask.distributed.Client, optional):
Distributed Client object for parallel parsing of anything that isn't a dask dataframe.
scheduler (dask.distributed.Client, optional)
Scheduler to use for parallellized parsing of dask dataframes.
can e.g. be simply the ``distributed.Client.get`` method.
Default is None.
dumper (module, optional):
Dumper to use for saving pandas dataframes.
Default is :py:mod:`~data_base.isf_data_base.IO.LoaderDumper.pandas_to_msgpack`.
.. deprecated:: 0.2.0
The :paramref:`burst_times` argument is deprecated and will be removed in a future version.
File: /gpfs/soma_fs/scratch/meulemeester/project_src/in_silico_framework/data_base/isf_data_base/db_initializers/load_simrun_general.py
Type: function
Configuring ISF¶
Dask¶
ISF works best with a Dask scheduler and workers for parallellization. If you are running the pixi installation (recommended), then you can simply start these with pixi:
pixi run launch_dask_server
and
pixi run launch_dask_workers
This will launch a server on the default port 8786, a dashboard on port 8787, accessible through your browser on http://localhost:8787. The default amount of workers is 4.
If you are not using the pixi installation, the above commands are simply shortcuts for the following two commands:
dask-scheduler --port=8786 --bokeh-port=8787 --host=localhost
and
dask-worker localhost:8786 --nthreads=1 --nprocs=4 --memory-limit=100e15 --dashboard-address=8787
If you are using ISF in a High-Performance Computing (HPC) context, you may want to adapt these default values.
After launching a Dask scheduler, you can connect to it from your notebook with a Dask Client. This client can then be used to execute code parallellized, and the dashboard will show the progress. ISF makes extensive use of Dask for parallellization, as seen in the example simulation below.
[4]:
dask_client = I.get_client()
dask_client
[4]:
Client
|
Cluster
|
Example simulation¶
Running a simulation from parameterfiles¶
As an example, let’s simulate a Layer 5 pyramidal cell in the rat barrel cortex under the in vivo condition of a single whisker deflection. We start by creating a DataBase for our simulation results. Feel free to adapt the path.
[5]:
from pathlib import Path
tutorial_output_dir = f"{Path.home()}/isf_tutorial_output" # <-- Change this to your desired output directory
db = I.DataBase(tutorial_output_dir).create_sub_db("intro_to_isf")
Now, let’s copy over some example parameter files. The tutorials provide more explanation on what these parameter files are, and how to create them. For now, we simply copy a pre-existing result.
[6]:
from getting_started import getting_started_dir
example_cell_parameter_file = I.os.path.join(
getting_started_dir,
'example_data',
'biophysical_constraints',
'86_C2_center.param')
example_network_parameter_file = I.os.path.join(
getting_started_dir,
'example_data',
'functional_constraints',
'network.param')
# Create new database entries
# If they already exist, they will NOT be overwritten, but also not raise an error
I.shutil.copy(example_cell_parameter_file, db.create_managed_folder('biophysical_constraints', raise_ = False))
I.shutil.copy(example_network_parameter_file, db.create_managed_folder('functional_constraints', raise_ = False))
example_cell_parameter_file = db['biophysical_constraints'].join('86_C2_center.param')
example_network_parameter_file = db['functional_constraints'].join('network.param')
[7]:
# Create new folder for results
db.create_managed_folder('simrun', raise_ = False)
dir_prefix = db['simrun'].join('stim_C2')
nSweeps = 1 # number of consecutive simulation trials per process
nprocs = 1 # number of processes simulating that task in parallel
tStop = 300 # stop the simulation at 300ms
d = I.simrun_run_new_simulations(
example_cell_parameter_file,
example_network_parameter_file,
dirPrefix = dir_prefix,
nSweeps = nSweeps,
nprocs = nprocs,
tStop = tStop,
silent = False)
[8]:
futures = dask_client.compute([d])
Use the dashboard to inspect the progress:
[9]:
dask_client
[9]:
Client
|
Cluster
|
Parsing simulation data¶
We use the data_base module through Interface to parse the data.
[10]:
I.db_init_simrun_general.init(
db,
db['simrun'],
client=dask_client
)
[WARNING] client: Connection dropped: socket connection broken
[WARNING] client: Transition to CONNECTING
[WARNING] client: Session has expired
Visualizing the simulation¶
We can create a Cell object from each simulation trial: a dataclass containing the simulation results.
[11]:
cell = I.simrun_simtrail_to_cell_object(
db,
sim_trial_index = db['sim_trial_index'][0]
)
[12]:
sa = db["synapse_activation"]
unique_syn_types = I.np.unique(sa["synapse_type"])
population_to_color_dict = {'L1': 'cyan', 'L2': 'dodgerblue', 'L34': 'blue', 'L4py': 'palegreen',
'L4sp': 'green', 'L4ss': 'lime', 'L5st': 'yellow', 'L5tt': 'orange',
'L6cc': 'indigo', 'L6ccinv': 'violet', 'L6ct': 'magenta', 'VPM': 'black',
'INH': 'grey', 'EXC': 'red', 'all': 'black', 'PSTH': 'blue'}
cmv = I.CellMorphologyVisualizer(
cell,
t_start=245-25,
t_stop=245+25,
t_step=0.2
)
for synapse_type in unique_syn_types:
celltype = synapse_type.split('_')[0]
if celltype not in population_to_color_dict: cmv.population_to_color_dict[synapse_type] = 'white'
else: cmv.population_to_color_dict[synapse_type] = population_to_color_dict[synapse_type.split('_')[0]]
cmv.population_to_color_dict["inactive"] = "#f0f0f0"
images_path = str(db._basedir/'C2_stim_animation_3d')
cmv.animation(
images_path=images_path,
color="voltage",
show_synapses=True,
client=dask_client,
)